
- 技術說明
- 論文資料
- Poster
什麼是SNAQTM-SEQ?
Standardized Nucleic Acid Quantification for SEQuencing(SNAQTM-SEQ) 為NGS定序時使用的標準化核酸定量方法,它利用在樣本萃取或 NGS library製備步驟之前將全合成製作的 DNA 或 RNA 內標準品 (Internal Standards) 加入到每個樣本中。內部標準品 (Internal Standards) 是監控定序結果變化的最佳方式,也是生物資訊分析的黃金標準!
實驗流程
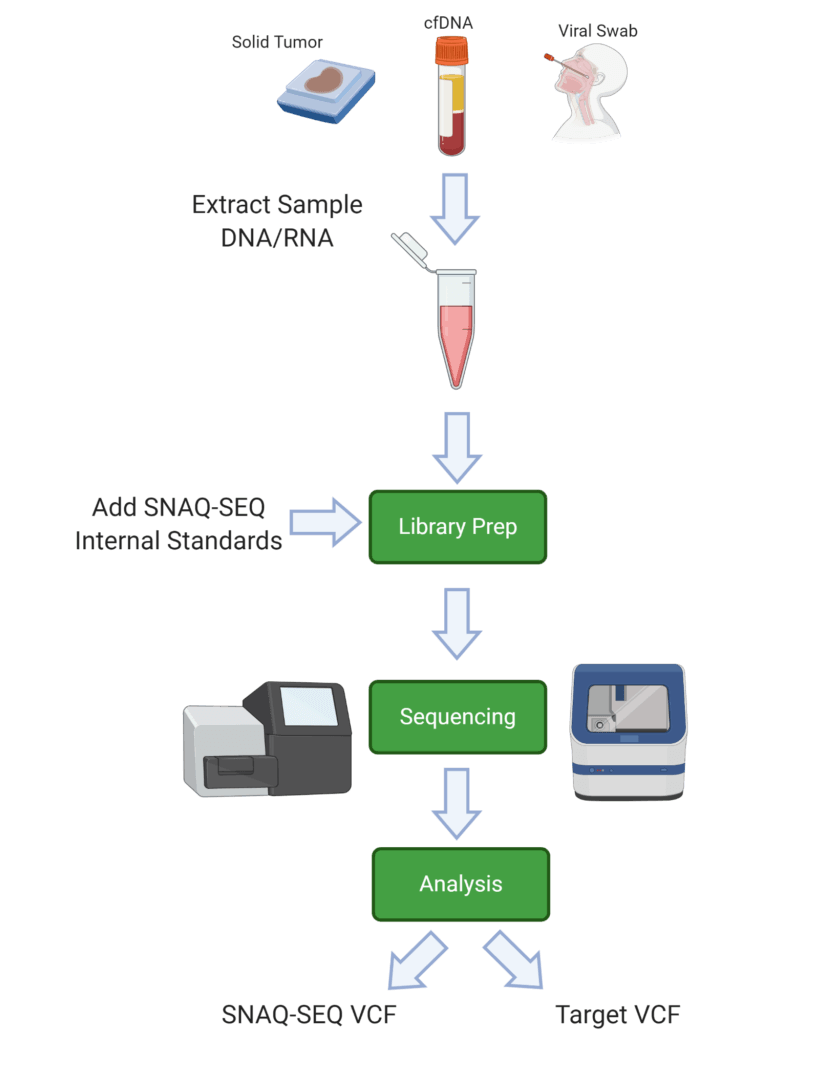
• 在Library preparation階段將內標準品(IS),添加入樣本中,
• IS與樣本一同反應與定序。
• 因IS序列中內建Barcode,於定序結果中可以辨識IS控制組和樣本的定序資料 。
為何需要內部標準品(Internal Standards)?
次世代定序已常用於臨床檢測、癌症篩檢與藥物研發等領域,但從樣本製備中可能造成的鹼基變化,致使 NGS 判讀錯誤,或臨床樣本量太低,無法精準分析等問題。 AccuGenomics利用全合成帶有獨特 Barcode的內標準品 (IS),可提升臨床檢體定序的靈敏度,並可對於每一個樣本進行內部QC檢測。
加入樣本中進行相同反應,為每個樣本提供獨立的QC標準
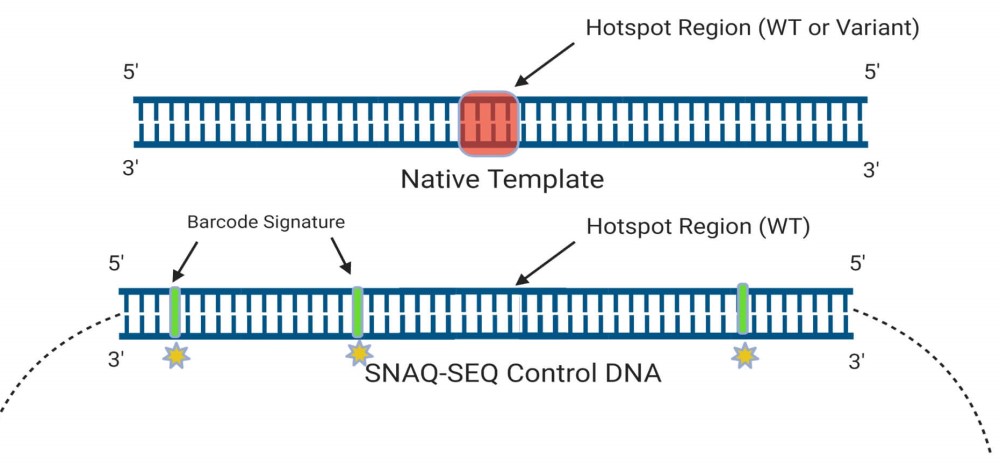
以全合成的方式製作目標區域的基因組序列, 可製作 DNA或 RNA片段,長度為60bp-2kb, 每隔50-80bp內建Barcode辨識,以區分標準 品與樣本的定序資料。
作為定序的黃金標準,排除操作流程內的任何系統性錯誤
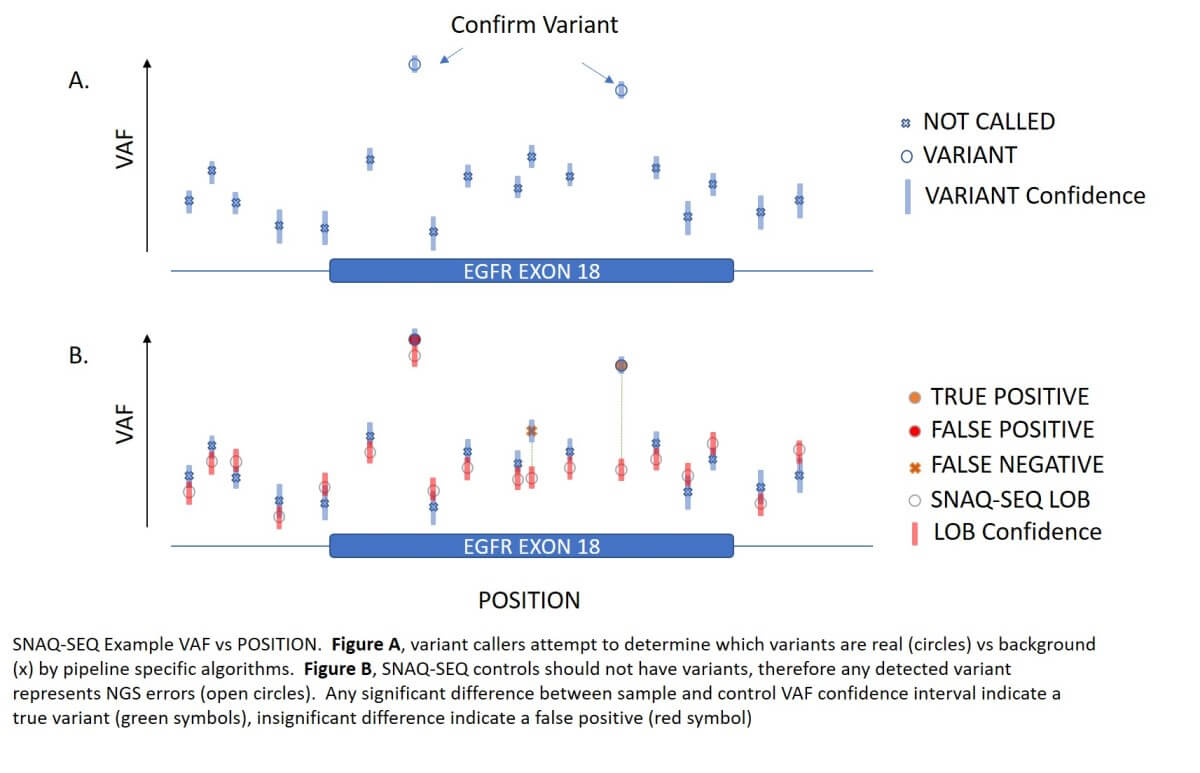
IS為每個樣本中的每個區域提供 Limit of Blank,提高變異檢出的準確性,幫助排除偽陽性和偽陰性,及系統或人為錯誤。適用於受到高背景值影響的低變異等位基因頻率(VAF)樣本。
利用加入的內標準品,可回推樣本的真實數據
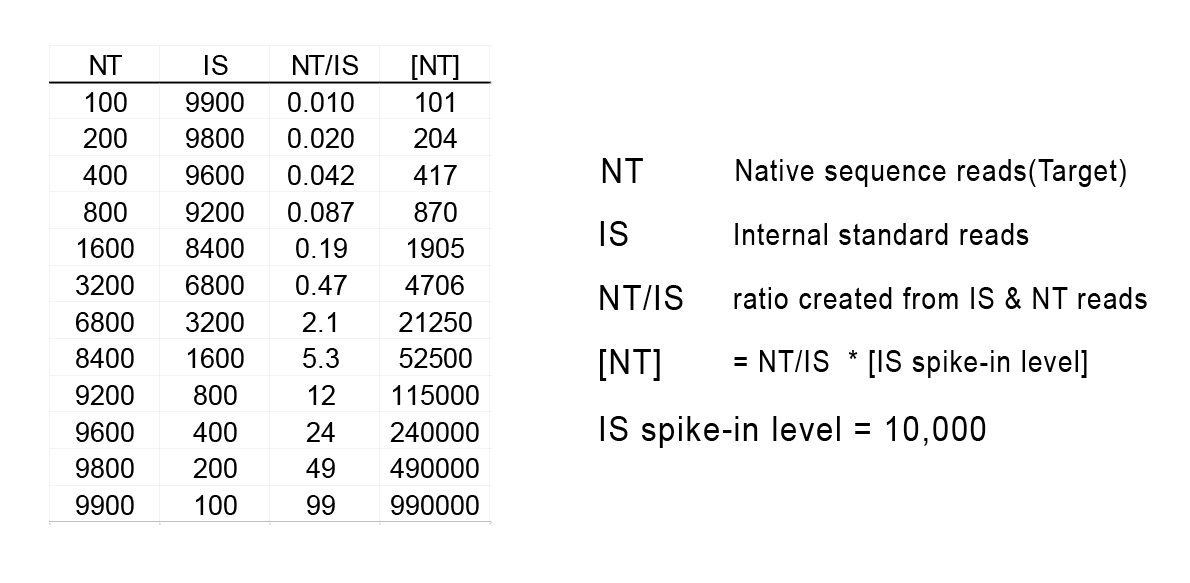
Standardized Nucleic Acid Quantification for SEQuencing(SNAQTM-SEQ) 為NGS定序時使用的標準化核酸定量方法,它利用在樣本萃取或 NGS library製備步驟之前將全合成製作的 DNA 或 RNA 內標準品(Internal Standards) 加入到每個樣本中。依照標準品與樣本的測序讀值比率,不須進行高深度定序, 即可達到如 dPCR 般的效果。
AccuGenomics 原廠說明文:
How to achieve Optimal SNAQ-SEQ Internal Standard Spike in Levels
Willey, J.C., Morrison, T.B., Austermiller, B., Crawford, E.L., Craig, D.J., etc.(2021). Advancing NGS quality control to enable measurement of actionable mutations in circulating tumor DNA. Cell Reports Methods
Gong, B., Li, D., Kusko, R., Novoradovskaya, N., Zhang, Y., Wang, S., Pabón-Peña, C., etc.(2021). Cross-oncopanel study reveals high sensitivity and accuracy with overall analytical performance depending on genomic regions. Genome Biology
Deveson, I.W., Gong, B., Lai, K., LoCoco, J.S., Richmond, T.A., Schageman, J., Zhang, Z., etc.(2021). Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nature Biotechnology
Craig, D. J., Morrison, T., Khuder, S. A., Crawford, E. L., Wu, L., Xu, J., Blomquist, T. M., & Willey, J. C. (2019). Technical advance in targeted NGS analysis enables identification of lung cancer risk-associated low frequency TP53, PIK3CA, and BRAF mutations in airway epithelial cells. BMC Cancer,
Craig, D.J., Blomquist, T.M., Crawford, E.L., Xu, J., Wu, L., Morrison, T. and Willey, J.C., (2019). Poster: Abstract #G009 Use of Synthetic Internal Standards to Measure Very Low Frequency TP53, PIK3CA, and BRAF.
Craig, D.J., Crawford, E.L., Xu, J., Blomquist, T.M., Wu, L., Morrison, T. and Willey, J.C., (2019). Poster: Novel method for NGS analysis of actionable mutations in circulating tumor DNA specimens: Improved quality control and 20-fold lower sequencing required
Morrison, T., Austermiller, B., Lazaridis, nick, Holshouser, C., Smith-Moore, C., O’Connell, K., Bernhardt, P., Fowler, V., Scott, M., Thomas, P. and Hoover, N., (2019). Poster: NIIMBL Adventitious Agent Detection by NGS. NIIMBL National Meeting 2019. Washington DC.
deAbreu, F., Deharvengt, S., Austermiller, B., Morrison, T., Tsongalis, G. and Lefferts, J., (2018). Poster: Spike in NGS Controls for Copy Number Assessment and Improved LOD and VAF Confidence. AMP 2018 Annual Meeting. San Antiono, Texas.
Willey, J.C., Blomquist, T.M., Crawford, E.L., Xu, J., Wu, L. and Morrison, T., (2018). Abstract 1623: Inter-laboratory harmonization of next generation sequencing somatic mutation assays for cancer response prediction. Clinical Research.
Yeo, J., Crawford, E. L., Zhang, X., Khuder, S., Chen, T., Levin, A., Blomquist, T. M., & Willey, J. C. (2017). A lung cancer risk classifier comprising genome maintenance genes measured in normal bronchial epithelial cells. BMC Cancer
Blomquist, T., Crawford, E. L., Yeo, J., Zhang, X., & Willey, J. C. (2015). Control for stochastic sampling variation and qualitative sequencing error in next generation sequencing. Biomolecular Detection and Quantification.
Yeo, J., Crawford, E.L., Blomquist, T.M., Stanoszek, L.M., Dannemiller, R.E., Zyrek, J., De Las Casas, L.E., Khuder, S.A. and Willey, J.C., (2014). A Multiplex Two-Color Real-Time PCR Method for Quality-Controlled Molecular Diagnostic Testing of FFPE Samples. PLoS ONE.
Blomquist, T. M., Crawford, E. L., Lovett, J. L., Yeo, J., Stanoszek, L. M., Levin, A., Li, J., Lu, M., Shi, L., Muldrew, K., & Willey, J. C. (2013). Targeted RNA-Sequencing with Competitive Multiplex- PCR Amplicon Libraries. PLoS ONE.

